Events Calendar
Yibing Shan
Tuesday, July 02, 2019, 02:30pm - 03:30pm
Hits : 3646
Contact Dima Kozakov, Host
Research Scientist, Chemistry, Biology and Drug Discovery
D. E. Shaw Research, NYC
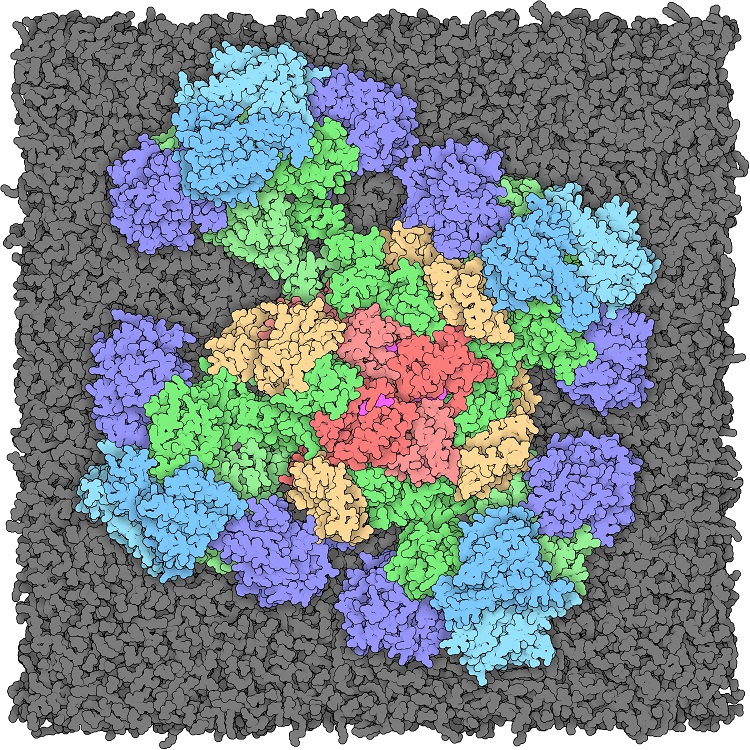
Structural modeling of large biomolecular assemblies--case studies on full-length JAK2 kinase and on Ras-Raf signalosome.
Molecular dynamics (MD) simulations hold the potential in recapitulating the physical process of protein-protein and protein-small molecule binding and in predicting the native structure of the biomolecular complexes. Careful scrutiny of the simulation-generated structural models in light of existing biochemical, biophysical, and cellular data should largely eliminate the false models and lead to focus on the correct ones. Further model constructions, MD simulations, and experimental validation enable may produce atomic-detailed structural models of large biomolecular assemblies. We here discuss two projects: we modeled the four-domained structures of inactive and active (dimerized) JAK2 kinase and the megadalton membrane-anchored Ras-Raf signalosome that consists of many copies of K-Ras and C-Raf proteins, together with copies of several auxiliary proteins. These models illustrate that MD simulation is increasing a viable platform for the structural biology of large biomolecular assemblies.

Location Laufer Center Lecture Hall 101
Refreshments following seminar in Laufer Hub 110


