Events Calendar
Senior Staff Scientist, Biological Nanostructures
The Molecular Foundry
Lawrence Berkeley National Laboratory
Berkeley, CA
How to build atomically defined nanostructures from sequence-defined peptoid polymers
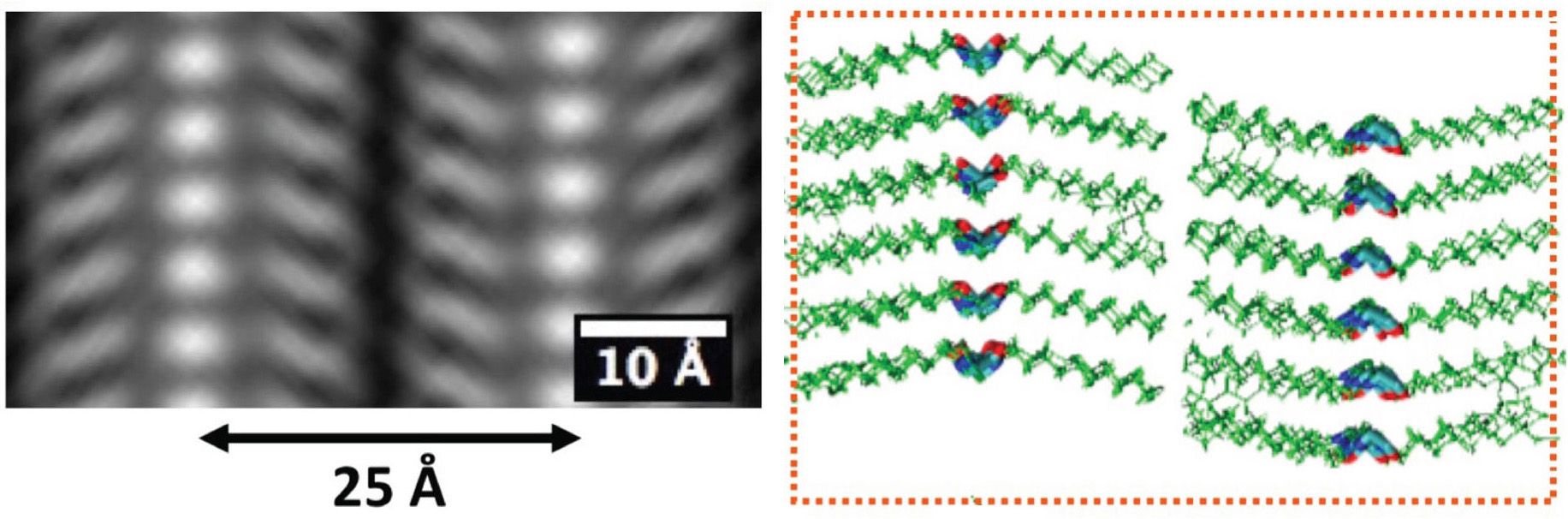
A fundamental challenge in materials science is to build synthetic nanostructures with the same architectural sophistication as proteins. One of the most promising ways to do this is to copy Nature, and synthesize sequence-defined, non-natural polymer chains that spontaneously fold and assemble into precise three-dimensional structures. To efficiently synthesize such a bio-inspired, information-rich material, we use the automated solid-phase submonomer synthesis method, which can readily generate high-purity, sequence-defined peptoid polymers up to 50 monomers in length. The method uses readily available primary amine synthons, allowing hundreds of chemically diverse sidechains to be cheaply introduced. We use this method, along with computational modeling, to design, synthesize, assemble and engineer a variety of protein-mimetic nanostructures, and to probe fundamental questions in polymer physics. Here we show by direct cryo-TEM imaging and x-ray scattering, that all known crystalline peptoid assemblies share a universal secondary structure motif based on a backbone fold containing all cis-amide bonds. This unexpected universality of peptoid backbone folding offers a unique opportunity to rationally design and engineer these materials to create robust, nanomaterials capable of protein-like functions, like specific molecular recognition and catalysis.