Events Calendar

Gabriel Rocklin, PhD
Assistant Professor
Department of Pharmacology
Northwestern University
Bio
Prof. Rocklin earned his Ph.D. in Biophysics at the University of California, San Francisco and then did postdoctoral research at the University of Washington’s Institute for Protein Design. He joined Northwestern’s Department of Pharmacology and Center for Synthetic Biology in 2019. His lab develops high-throughput experimental methods to understand protein biophysics and improve computational protein design. His lab is also a member of the international Rosetta Commons consortium for biomolecular modeling and design. In 2020, his lab was awarded an NIH New Innovator award to support their research into protein conformational dynamics.
High-throughput discovery of protein folding stability and dynamics
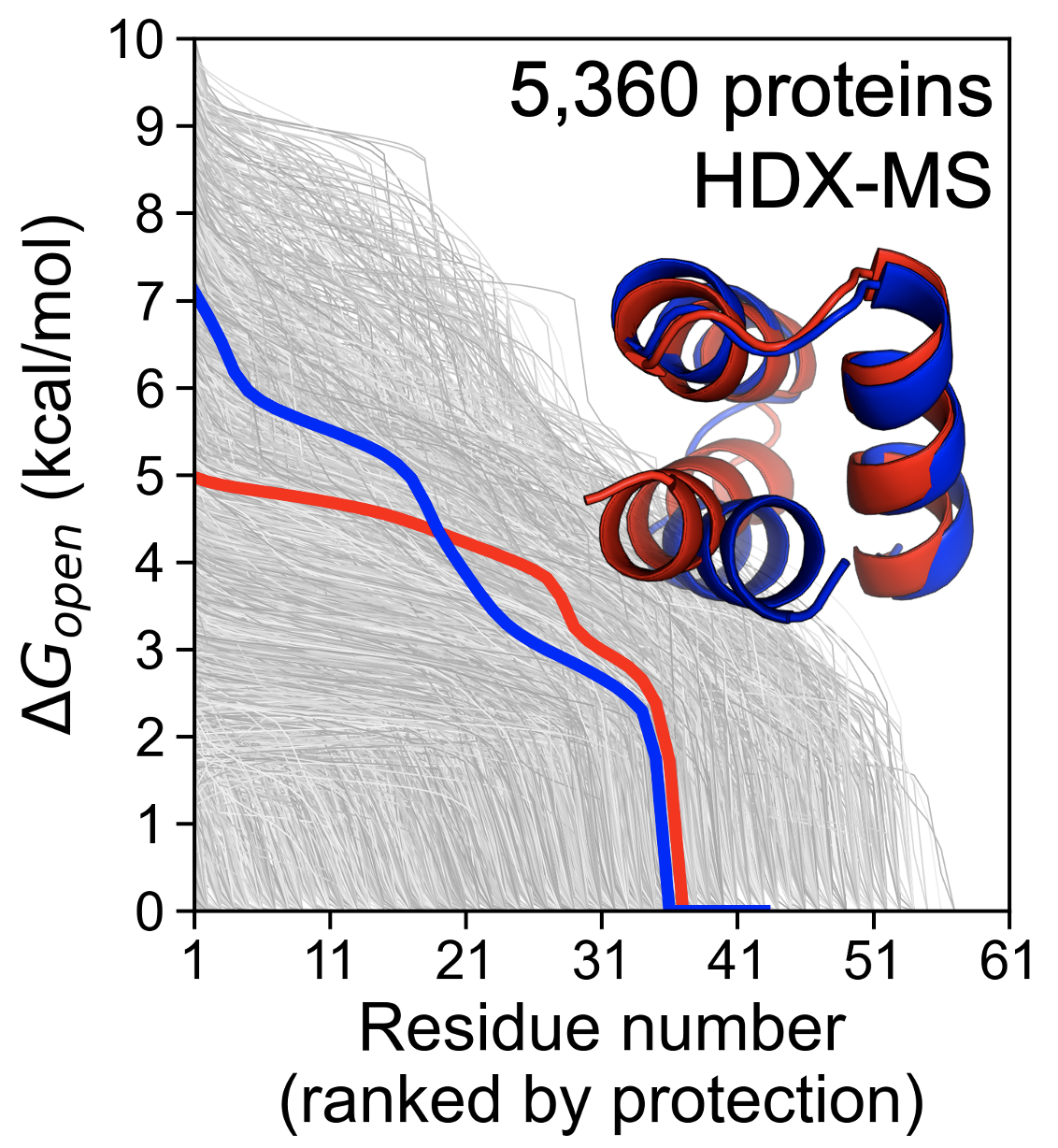
Protein sequences vary widely in folding stability, and optimizing stability remains a key challenge in protein engineering. Protein sequences also vary in their conformational dynamics: some sequences are "locked" into one rigid structure while others sample a landscape of diverse folded and partially-folded conformations. Machine learning methods promise to empower protein engineers to precisely tune stability and dynamics for specific applications, ushering in a new era of protein design. However, developing accurate tools is bottlenecked by the limited availability of experimental data measuring protein stability and dynamics. Our lab has pioneered high-throughput methods to quantify stability and dynamics on unprecedented scales, including recently released stability measurements for >750,000 protein domains and unpublished measurements quantifying conformational dynamics for >5,000 domains. I will share our lab's approach to developing these high-throughput assays as well as our findings on how protein sequences determine stability and dynamics. Furthermore, by comparing our stability measurements to our independent dynamics measurements, I will illustrate the complementarity between these approaches and the unique insights gained by pursuing both. Finally, I will share progress in developing predictive machine learning tools based on these new high-throughput data.
Host: Eugene Serebryany
Location: Laufer Center Lecture Hall 101
Refreshments following the seminar in room 110







